Chromosomes and Genes
Category : NEET
Chromosomes and Genes
The chromosomes are capable of self-reproduction and maintaining morphological and physiological properties through successive generations. They are capable of transmitting the contained hereditary material to the next generation. Hence these are known as ‘hereditary vehicles’. The eukaryotic chromosomes occurs in the nucleus and in certain other organelles, and are respectively called nuclear and extranuclear chromosomes. Nuclear chromosomes are long, double stranded DNA molecules of linear form and associated with proteins, separated from the cytoplasm by nuclear envelope and replicated during S phase of cell cycle, while extranuclear chromosomes are present in the mitochondria and plastid. They are short, double stranded DNA molecules of circular form and are not associated with proteins and also called prochromosomes.
(i) Discovery of chromosomes
Hofmeister (1848): First observed chromosomes in microsporocytes (microspore mother cells) of Tradescantia.
Flemming (1879): Observed splitting of chromosomes during cell division and coined the term, ‘chromatin’.
Roux (1883): He believed the chromosomes take part in inheritance.
W.Waldeyer (1888): He coined the term ‘chromosome’.
Benden and Boveri (1887): They found a fixed number of chromosomes in each species.
(ii) Kinds of chromosomes
(a) Viral chromosomes: In viruses and bacteriophages a single molecule of DNA or RNA represents the viral chromosome.
(b) Bacterial chromosomes: In bacteria and cyanobacteria, the hereditary matter is organized into a single large, circular molecule of double stranded DNA, which is loosely packed in the nuclear zone. It is known as bacterial chromosome or nucleoid.
(c) Eukaryotic chromosomes: Chromosomes of eukaryotic cells are specific individualized bodies, formed of deoxyribonucleo proteins (DNA + Proteins).
(iii) Number of chromosomes: The number of chromosomes varies from two, the least number an organism can have, to a few hundred in different species. The number of chromosomes a species possesses has no basic significance, nor it necessarily shows relationship between two different species that have the same number.
Both dog and fowl have 78 chromosomes. Thus, it is not the number of chromosomes, but the genes in them which differentiate species. Their number also does not indicate the size or complexity of the organism. Amoeba proteus has 250 chromosomes and man has 46. The related species tend to have similar chromosome. Man and his nearest relatives, the apes, have chromosomes similar in size, shape and banding pattern. The least number of chromosomes are found in Ascaris megalocephala i.e. 2 while in a radiolarian protist (Aulocantha) has maximum number of chromosomes is 1600. The male of some roundworms and insects have one chromosome less than the females. For instance, the male and female roundworm Coenorhabditis have 11 and 12 chromosomes respectively and the male and female cockroach (Blatta) have 23 and 24 chromosomes respectively.
|
Common name |
Zoological name |
Chromosomes |
|
(1) Man |
Homo sapiens |
46 |
|
(2) Gorilla |
Maccaca mulatta |
48 |
|
(3) Pig |
Sas scrofa |
40 |
|
(4) Sheep |
Ovis aries |
54 |
|
(5) Cat |
Felis maniculata |
38 |
|
(6) Dog |
Canis familiaris |
78 |
|
(7) Rat |
Rattus rattus |
42 |
|
(8) Rabbit |
Oryctolagus cuniculus |
44 |
|
(9) Honey bee |
Apis mellifera |
32, 16 |
|
(10) Mosquito |
Culex sp |
6 |
|
(11) Grasshopper |
Gryllus |
23(male), 24(female) |
(iv) Chromosome structure: Different regions or structure recognized in chromosomes are as under
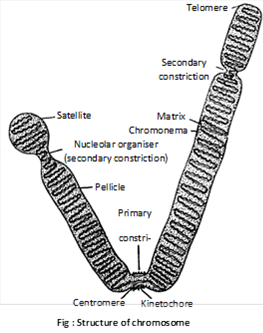
(a) Pellicle: It is the outer thin but doubtful covering or sheath of the chromosome.
(b) Matrix: Matrix or ground substance of the chromosome is made up of proteins, small quantities of RNA and lipid. It has one or two chromonemata (singular-chromonema) depending upon the state of chromosome.
(c) Chromonemata: They are coiled threads which form the bulk of chromosomes. A chromosome may have one (anaphase) or two (prophase and metaphase) chromonemata. The coiled filament was called chromonema by Vejdovsky in 1912. The coils may be of the following 2 types
(1) Paranemic coils: When the chromonemal threads are easily separable from their coils then such coils are known as paranemic coils.
(2) Plectonemic coils: When the chromosomal threads remain inter-twined so intimately that they cannot be separated easily are known as plectonemic coils.
(d) A primary Constriction and Centromere (kinetochore): A part of the chromosome is marked by a constriction. It is comparatively narrow than the remaining chromosome. It is known as primary constriction. The primary constriction divides the chromosome into two arms. It shows a faintly positive Feulgen reaction, indicating presence of DNA or repetitive type. This DNA is called centromeric heterochromatin.
Centromere or kinetochore lies in the region of primary constriction. The microtubules of the chromosomal spindle fibres are attached to the centromere. Therefore, centromere is associated with the chromosomal movement during cell division. Kinetochore is the outermost covering of centromere.
Type of chromosomes based on number of centromeres: Depending upon the number of centromeres, the chromosomes may be:
(1) Monocentric with one centromere.
(2) Dicentric with two centromeres, one in each chromatid.
(3) Polycentric with more than two centromeres.
(4) Acentric without centromere. Such chromosomes represent freshly broken segments of chromosomes, which do not survive for long.
(5) Diffused or non-located with indistinct throughout the length of chromosome. The microtubules of spindle fibres are attached to chromosome arms at many points. The diffused centromeres are found in insects, some algae and some groups of plants.
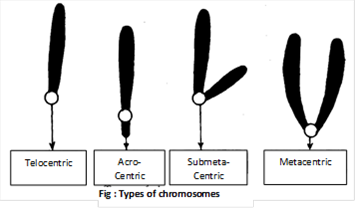
Types of chromosomes based on position of centromere: Based on the location of centromere the chromosomes are categorised as follows:
(1) Telocentric: These are rod-shaped chromosomes with centromere occupying a terminal position. One arm is very long and the other is absent.
(2) Acrocentric: These are rod-shaped chromosomes having subterminal centromere. One arm is very long and the other is very small.
(3) Submetacentric: These are J or L shaped chromosomes with centromere slightly away from the mid-point so that the two arms are unequal.
(4) Metacentric: These are V-shaped chromosomes in which centromere lies in the middle of chromosomes so that the two arms are almost equal.
(e) Chromomeres: Chromomeres are linearly arranged bead-like and compact segments described by J. Bellings. They are identified by their characteristic size and linear arrangement along a chromosome.
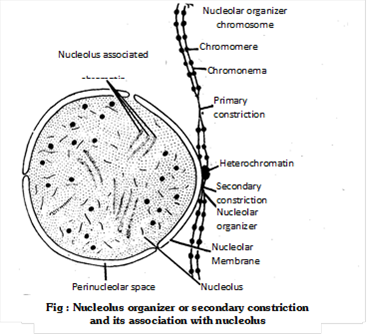
(f) Secondary constriction or nucleolar organizer: Sometimes one or both the arms of a chromosome are marked by a constriction other than the primary constriction. During interphase this area is associated with the nucleolus and is found to participate in the formation of nucleolus. It is, therefore, known as nucleolar organizer region or the secondary constriction. In certain chromosomes, the secondary constriction is (In human beings 13, 14, 15, 20 and 21 chromosome are nucleolar organizer) intimately associated with the nucleolus during interphase. It contains genes coding for 18S and 28S ribosomal RNA and is responsible for the formation of nucleolus. Therefore, it is known as nucleolar organizer region (NOR).
(g) Telomeres: The tips of the chromosomes are rounded and sealed and are called telomeres which play role in Biological clock. The terminal part of a chromosome beyond secondary constriction is called satellite. The chromosome with satellite is known as sat chromosome, which have repeated base sequence.
(h) Chromatids: At metaphase stage a chromosome consists of two chromatids joined at the common centromere. In the beginning of anaphase when centromere divides, the two chromatids acquire independent centromere and each one changes into a chromosome.
(v) Molecular organisation of chromosome: Broadly speaking there are two types of models stating the relative position of DNA and proteins in the chromosomes.
(a) Multiple strand models: According to several workers (Steffensen 1952, Ris 1960) a chromosome is thought to be composed of several DNA protein fibrils and atleast two chromatids form the chromosome.
(b) Single strand models: According to Taylor, Duprow etc. The chromosome is made up of a single DNA protein fibril. There are some popular single strand models.
(1) Folded fibre model: Chromosomes are made up of very fine fibrils 2 nm - 4 nm in thickness. As the diameter of DNA molecule is also 2 nm (\[20{\AA}\]). So it is considered that a single fibril is a DNA molecule. It is also seen that chromosome is about a hundred times thicker than DNA whereas the length of DNA in chromosome is several hundred times that of the length of chromosome. So it is considered that long DNA molecule is present in folding manner which forms a famous model of chromosome called folded fibre model which given by E.J. Dupraw (1965).
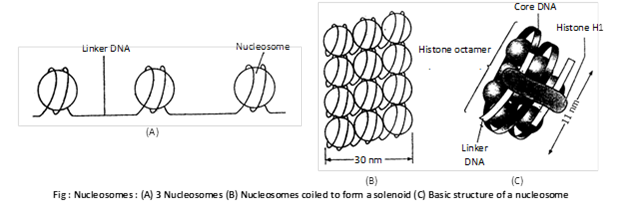
(2) Nucleosome model: The most accepted model of chromosome or chromatin structure is the ‘nucleosome model’ proposed by Kornberg and Thomas (1974). Nucleosomes are also called core particles or Nu-bodies. The name nucleosome was given by P. Outdet et al. The nucleosome is a oblate particle of \[55{\AA}\] height and \[110{\AA}\] diameter. Woodcock (1973) observed the structure of chromatin under electron microscope. He termed each beaded structure on chromosome as nucleosome. Nucleosome is quasicylindrical structure made up of histones and DNA.
Histone are mainly of two types:
(a) Nucleosomal histone: These are small proteins responsible for coiling DNA into nucleosome. These are\[{{H}_{2}}A\],\[~{{H}_{2}}B,{{H}_{3}}and{{H}_{4}}\]. Each histone protein consist of two molecule, thus the four histone proteins form a octamer. These form the inner core of nucleosome.
(b) Linker histone: \[{{H}_{1}}\] proteins is known as linker histone that connect one core particle with another. These are present once per 200 base pairs. These are loosely associated with DNA. \[{{H}_{1}}\] histone are responsible for packing of nucleosome into 30 nm fibre.
Functions of histones: Histones in eukaryotic chromosomes serve some functions.
DNA in nucleosome: Nucleosome is made of core of eight molecules of histones wrapped by double helical DNA with \[1\frac{3}{4}\] turns making a repeating unit. Every \[1\frac{3}{4}\] turn of DNA have 146 base pairs. When \[{{H}_{1}}\]protein is added the nucleotide number becomes 200. DNA which joins two nucleosome is called linker DNA or spacer DNA.
(3) Solenoid model: In this model the nucleosomal bead represents the first degree of coiling of DNA. It is further coiled to form a structure called solenoid (having six nucleosome per turn). It represents the second degree of coiling. The diameter of solenoid is \[300{\AA}\]. The solenoid is further coiled to form a supersolenoid of \[2000-4000{\AA}\] diameter. This represent the third degree of coiling. The supersolenoid is perhaps the unit fibre or chromonema identified under light microscopy. The solenoid model was given by Fincy and Klug 1976. Klug was awarded by nobel prize in 1982 for his work on chromosome.
(4) Dangier-String or Radial Loop Model: (Laemmli, 1977). Each chromosome has one or two interconnected scaffolds made of nonhistone chromosomal proteins. The scaffold bears a large number of lateral loops all over it. Both exit and entry of a lateral loop lie near each other. Each lateral loop is 30 nm thick fibre similar to chromatin fibre. It develops through solenoid coiling of nucleosome chain with about six nucleosomes per turn. The loops undergo folding during compaction of chromatin to form chromosome.
(vi) Heterochromatin and Euchromatin: Flemming (1880) named the readily stainable material in nuclei as chromatin. It is present both during interphase and cell division (as the chromosomal material). It consists of about equal parts by weight of DNA and histones. There are two classes of chromatin structure, heterochromatin and euchromatin.
(a) Heterochromatin or static chromatin is highly condensed and is usually transcriptionally inactive and found in the centromeres of chromosomes. Heterochromatin is of two types, (i) genetically inactive constitutive heterochromatin which is a permanent part of the genome, and (ii) facultative heterochromatin which varies in its state in different cell types and development stages.
(b) Euchromatin or dynamic chromatin is relatively extended and open. It at least has the potential of being actively transcribed. It makes up the major part of the genome, and is visible only during mitosis.
Comparison of heterochromatin and euchromatin
|
Heterochromatin |
Euchromatin |
|
Remains condensed throughout interphase (positive heteropycnosis) giving rise to chromocentres. |
Shows normal cycle of condensation during cell division and extension during interphase. |
|
Because of the condensed state, it stains more heavily giving rise to banding patterns or chromosomes. |
Because it is less condensed, it stains less heavily (normal staining properties). Only slightly basophilic. |
|
Found in condensed regions of the chromosome and in association with tight folding or coiling of the chromosomal fibre. |
Found diffuse or less tightly coiled regions. Undergoes typical condensation-decondensation cycle. |
|
May contain highly repetitive (satellite) DNA or single copy (unique) DNA. |
Almost free of repetitive DNA. Contains predominantly single copy DNA. |
|
Relatively inert metabolically, but does not contain a few genes. |
Genetically active: Almost all the genes are located on euchromatin. |
|
DNA is genetically inert, and does not transcribe mRNA for protein synthesis in the condensed state. |
Genetically active, dispersed part of chromatin in interphase nuclei. Its DNA synthesizes mRNA for protein synthesis. |
|
Late replication of DNA at the S phase of the cell cycle. Under-replication in polytene chromosomes. |
Comparatively early replication of DNA during the early stage of the S phase of cell cycle. |
|
Crossover frequency is less, because condensed regions of the chromosomal fibre cannot come close together for frequent crossover. This may help protect vital genes from the effects of crossover. |
Crossover frequency is more because of the decondensed (extended) state of euchromatin. |
(vii) Chromosome banding : It was the technique demonstrated by Casperson (1968) using a fluorescent dye quinacrine mustard for the study of finer chromosomal aberrations. The development of banding techniques has made the identification of individual chromosomes easier. Each chromosome can be identified by its characteristic banding pattern. In X chromosomes the bands are large, each containing \[\tilde{\ }{{10}^{7}}bp\] of DNA, and could include several hundreds of genes. The different banding techniques are identified by the letters Q, G, C, R and T.
Type of banding |
Staining technique |
Nature of bands |
|
Q (quinacrine) banding |
Chromosomes exposed to quinacrine mustard (acridine dye) which preferentially binds to AT-rich DNA. Other fluorescent dyes used are DAPI or Hoeschst 33258. |
UV fluorescence reveals fluorescencing Q bands which correspond to G-bands. DNA of Q/G bands contains more closely spaced SARs, giving tighter loops (Q loops). |
|
G (Giemsa) banding |
Chromosomes treated with alkaline solution and subjected to controlled trypsin digestion before staining with Geimsa, a DNA banding chemical dye. Relatively permanent stain. |
Dark bands are called G bands and pale bands are G-negative. G bands are presumed to be AT-rich. They are late replicating and contain highly condensed chromatin. |
|
R (reverse) banding |
Chromosomes treated with heated saline or restrictase to denature AT-rich DNA and stained with Giemsa. GC-specific chromomycin dyes, e.g, chromomycin A, olivomycin or mithracin give the same pattern. |
R-banding pattern is essentially the reverse of the G-banding pattern. R bands are Q negative. They generally replicate in the S-phase and have less condensed chromatin. |
|
T (telomaric) banding |
Prolonged heat treatment of chromosomes before staining with Giemsa or combination of dyes and fluorochromes. |
T bands are a subset of R bands which are the most inetnsely staining. They are especially concentrated at the telomeres. |
|
C (centromere) banding |
Chromosomes pre-treated with sodium hydroxide or barium hydroxide and stained with Giemsa. |
Preferred darkening of constitutive centromeric heterochromatin. Rest of the chromosome show Q banding pattern. |
(viii) Human karyotype and idiogram: Tjio and Levan (1956) of Sweden found that human cells have 23 pairs or 46 chromosomes. 22 pairs or 44 chromosomes are autosome and the last or 23rd pairs is that of sex chromosomes, XX in females and XY in males. A set of chromosomes of an individual or species is called a karyotype. In human the 23 pairs of chromosomes in somatic cells form the karyotype. It is possible to identify individual chromosomes on the basis of the following characteristics.
(1) The total length of the chromosomes.
(2) Arm ratio.
(3) The position of the secondary constrictions and nucleolar organizers.
(4) Subdivision of the chromosome into euchromatic and heterochromatic regions.
Homologous pairs of identified chromosomes can be arranged in a series of decreasing lengths. Such an arrangement is called an idiogram. Idiogram not possible in symmetrical karyotype.
(a) Karyotyping of human chromosomes: Chromosomes are clearly visible only in rapidly dividing cells. Human chromosomes are studied in blood cells (WBCs), cells in bone marrow, amniotic fluid and cancerous tissues. The WBCs divide when added with phytohaemagglutinin (PHA). The division stops when colchicine is added at metaphase stage. These dividing WBCs are then treated with hypotonic saline solution. Chromosomes are now stained with stains like orcein, Giemsa dye or recent quinacrine dye. When viewed with special miscroscope in ultraviolet light the stain produces fluorescent bands on chromosomes. The chromosomes are then arranged on photographic plate for making diagram and their study. The pictorial representation of a person’s chromosomes is called Karyotype.
(b) Classification of chromosomes: The human metaphase chromosomes were first of all classified by a conference of cytogeneticists at Denver, Colorado in 1960 and is known as the 23 pairs (46) chromosomes in human has been numbered from 1 to 23 according to their decreasing size. Patau (1960) divided the human chromosome into the following seven groups designated A to G.
|
Group |
Size |
Shape |
Number in set |
Number in a cell |
|
A |
Large |
Metacentric Submetacentric |
1-3 |
6 |
|
B |
Large |
Submetacentric |
4-5 |
4 |
|
C |
Medium |
Submetacentric |
6-12 and X |
15 male 16 female |
|
D |
Medium |
Acrocentric |
13-15 |
6 |
|
E |
Small |
Submetacentric |
16-18 |
6 |
|
F |
Small |
Metacentric |
19-20 |
4 |
|
G |
Smallest |
Acrocentric |
21-22 and Y |
5 male 4 female |
|
46 |

(ix) Special types of chromosomes
(a) Supernumerary, Accessory or B chromosomes or Satellite chromosomes or Giant lines plasmid: In some species, chromosomes have been found that are in addition to the normal autosomes and heterosomes. These chromosomes have been called supernumerary chromosomes, accessory chromosomes or B-chromosomes, and differ from normal or A-chromosomes in the following respects.

(1) They are usually smaller than A-chromosomes.
(2) They are frequently heterochromatic and telocentric.
(3) They are genetically unnecessary, and normally do not strongly influence viability and phenotype.
(4) Their number may vary in different cells, tissues, individuals and populations.
(5) They are not homologous with any of the A-chromosomes and do not synapses with them.
(6) They are found more commonly in plants than in animals.
Among animals they have been reported mostly in insects and a few species of flatworms. Of the 50 species of insect in which B-chromosomes have been reported, 29 are short horned grasshoppers belonging to the family Acridiae. Usually each nucleus has one or two B-chromosomes. In Tradescantia edwardsiana there are 50 B-chromosomes in addition to the 12 somatic A-chromosomes.
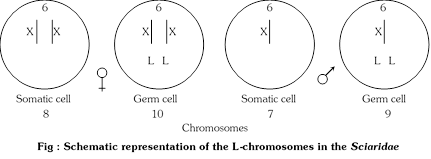
(b) Limited or L-chromosomes: Limited or L-chromosomes are so called because they are limited to the germ line. They have been found in the family Sciaridae (Diptera: Insecta). The germ line cells in females have 10 chromosomes: three pairs of autosomes, a pair of X-chromosomes and a pair of L-chromosomes. Those of males have 9 chromosomes, there being only one X-chromosome. Somatic cells have 8 chromosomes in females and 7 in males, the L-chromosomes being absent. During the fifth and sixth cleavages, L-chromosomes are eliminated from nuclei destined to form somatic tissue, but retained in germ line cells. L-chromosomes differ from B-chromosomes in that they are constant in all individuals of the species having them. B-chromosomes are found only in some individuals of the species.
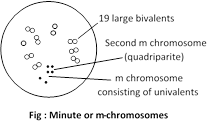
(c) Minute or m-chromosomes: Minute or m-chromosomes are so called because of their extremely small size (0.5 micron or less). They have been found in a variety of species of bryophytes, higher plants, insects of the family Coreidae (Heteroptera) and birds. They have been seen mainly during meiosis, and only occasionally during mitosis. Usually one or two chromosomes are seen, but four to five may also be present. In the moss, Sphagnum there are 19 large bivalents and 2 m- chromosomes.
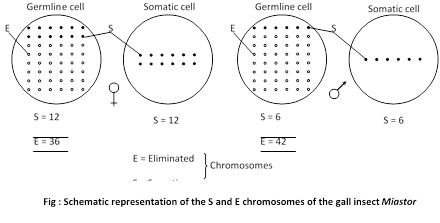
(d) S and E-chromosomes: S and E-chromosomes have been reported in insects in the family Cecidomyiidae (gall insects) and family Chironomidae (Diptera). In the gall insect Miastor, both males and females have 48 chromosomes in germ line cells. In somatic cells, however, there are only 12 chromosomes in females and 6 in males. Chromosomes which are present in both germ and somatic cells are called S-chromosomes. Those which are eliminated from somatic cells but are present in germ cells are called E-chromosomes. Thus in females the germ line cells have 12 S-chromosomes and 36 E-chromosomes. In male germ line cells there are 6 S-chromosomes and 42 E-chromosomes. The zygote receives half its S-chromosomes from each parent, while all the E-chromosomes are received from the female parent.
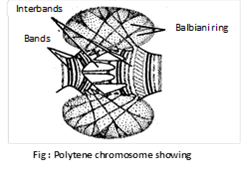
(e) Polytene chromosome: Polytene chromosome was described by Kollar (1882) and first reported by Balbiani (1881) in the salivary gland cells of chironomus larva. They are found in salivary glands of insects (Drosophila) and called as salivary gland chromosomes. These are reported in endosperm cells of embryosac by Malik and Singh (1979). Length of this chromosome may be upto 2000mm. The chromosome is formed by somatic pairs between homologous chromosomes and repeated replication or endomitosis of chromonemata. These are attached to chromocentre. It has pericentromeric heterochromatin. Polytene chromosomes show a large number of various sized intensity bands when stained. The lighter area between dark bands are called interbands. They have puffs bearing Balbiani rings. Balbani rings produce a number of m-RNA, which may remain stored temporarily in the puffs, are temporary structures. These are also occur in Malpighian tubules, rectum, gut, foot pads, fat bodies, ovarian nurse cells etc.
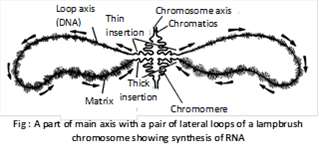
(f) Lampbrush chromosomes: They are very much elongated special type of synapsed or diplotene chromosome bivalents already undergone crossing over and first observed by Flemming (1882). The structure of lampbrush chromosome was described by Ruckert (1892). The lampbrush chromosomes occur at the diplotene stage of meiotic prophase in the primary oocytes of all animal species, both vertebrates and invertebrates as in sagitta (chaetognatha), sepia (mollusca), Echinaster (Echinodermata) and in several species of insect, sharks, amphibians, reptiles, birds and mammals. Lampbrush chromosomes are also found in spermatocytes of several species, giant nucleus of acetabularia and even in plants. In urodele oocyte the length of lampbrush chromosome is upto 5900mm. These are found in pairs consisting of homologous chromosomes jointed at chiasmata (meiotic prophase-I). The chromosome has double main axis due to two elongated chromatids. Each chromosome has rows of large number of chromatid giving out lateral loops, which are uncoiled parts of chromomere with one-many transcriptional units and are involved in rapid transcription of mRNA meant for synthesis of yolk and other substances required for growth and development of meiocytes. Some mRNA produced by lampbrush chromosome is also stored as informosomes i.e., mRNA coated by protein for producing biochemicals during the early development of embryo. Length of loop may vary between 5-100mm.
Important Tips
Genes Expression and Its Regulation
(1) Gene expression in prokaryotes: Gene expression refers to the molecular mechainism by which a gene expresses a phenotype by synthesizing a protein or an enzyme. Which determines the character. The gene contains the blue print or the information for the protein or an enzyme.
The category includes mechanism involved in the rapid turn-on and turn-off gene expression in response to environmental changes. Regulatory mechanism of this type are very important in microorganisms, because of the frequent exposure of these organisms to sudden changes in environment.
Gene concept can be studied by operon model. Operon are segment of genetic material which function as regulated unit that can be switched on and switched off, which was given by French scientists. Jacob and Monod (1961) working at Pasteur institute. They were studying lactose utilisation in mutants of E.coli. An operon consists of one to several structural genes (three in lac operon and five in tryptophan operon of Escherichia coli, nine in histidine operon of Salmonella typhimurium), an operator gene a promoter gene a regulator gene, a repressor and inducer or corepressor. Operons are of two types, inducible and repressible.
(i) Inducible operon system /lac operon system: An inducible operon system is that regulated genetic material which remains switched off normally but becomes operational in the presence of an inducer. It occurs in catabolic pathways. The components are:-
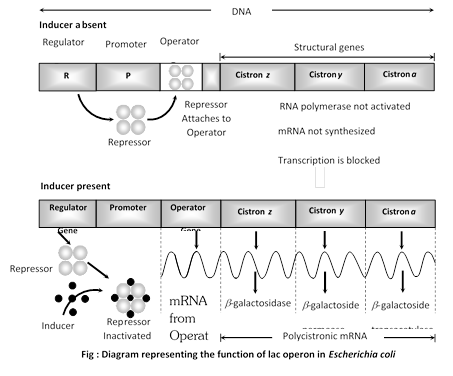
(a) Structural genes: They are genes, which produce mRNAs for forming polypeptides/proteins/enzymes. Lac operon of Escherichia coli has three structural genes-Z (produces enzyme \[\beta \]-galactosidase for splitting lactose/galactoside in to glucose and galactose) Y (produces enzyme galactoside permease required in entry of lactose/galactoside) and A (produces enzyme galactoside acetylase/transacetylase without any function in E.coli). The three structural genes of lac operon produce a single polycistronic mRNA. The three enzymes are, however, produced in different concentration.
(b) Operator gene: It gives passage to RNA polymerase when the structural genes are to express themselves. Normally, it is covered by a repressor. Operator gene of lac operon is small, made of 27 base pairs.
(c) Promoter gene: It is recognition centre / initiation point for RNA polymerase of the operon.
(d) Regulator gene (i Gene): It produces a repressor that binds to operator gene for keeping it nonfunctional (preventing RNA polymerase to pass from promoter to structural genes).
(e) Represso : It is a small protein formed by regulator gene. Which binds to operator gene and blocks passage of RNA polymerase towards structural enzymes. Repressor has two allosteric sites, one for attaching to operator gene and second for binding to inducer. Repressor of lac operon has a molecular weight of 160,000 and 4 subunit of 40,000 each.
(f) Inducer: It is a chemical which attaches to repressor, changes the shape of operator binding site so that repressor no more remain attached to operator.
Lactose/galctoside is inducer of lac operon. As soon as the operator gene becomes free, RNA polymerase is recognised by promoter gene. cAMP is required, RNA polymerase passes over the operator gene and then reaches the area of structural genes. Here it catalyses transcription of mRNAs.
|
Induction |
Repression |
|
It turns the operon on. |
It turns the operon off. |
|
It starts transcription and translation. |
It stops transcription and translation. |
|
It is caused by a new metabolite which needs enzymes to get metabolised. |
It is caused by an excess of existing metabolite |
|
It operates in a catabolic pathway. |
It operates in an anabolic pathway. |
|
Repressor is prevented by the inducer from joining the operator gene. |
Aporpressor is enabled by a corepressor to join the operator gene |
(ii) Repressible operon system/tryptophan operon system: A repressible operon system is that regulated genetic material, which normally remains active/operational and enzymes formed by its structural genes present in the cell till the operon is switched off when concentration of an end product crosses a threshold value. Repressible operon system usually occurs in anabolic pathways, e.g., tryptophan operon, argnine operon. Each has the following parts.
(a) Structural genes: They are genes, which take part in synthesis of polypeptides/proteins/enzymes through the formation of specific mRNAs. Tryptophan operon has five structural genes - E, D, C, B and A.
(b) Operator gene: It provides passage to RNA polymerase moving from promoter to structural genes. Operator gene of repressible operon is normally kept switched on as aporepressor formad by regulator gene is unable to block the gene.
(c) Promoter gene: It is initiation/recognition point for RNA polymerase.
(d) Regulator gene: The gene produces an aporepressor.
(e) Aporepressor: It is a proteinaceous substance formed through the activity of regulator gene. It is able to block operator gene only when a corepressor is also available.
(f) Corepressor: The nonproteinaceous component of repressor, which can be end product (feedback inhibition/repression) of the reaction mediated through enzymes synthesized by structural genes. Corepressor of tryptophan operon is tryptophan. It combines with aporepressor, form repressor which then blocks the operator gene to switch off the operon.
(2) Gene expression in eukaryotes: In regulation of gene expression in eukaryotes the chromosomal proteins play important role. The chromosomal proteins are of two types. They are histones and non-histones. The regulation of gene expression involves an interaction between histones and non-histones. Histones inhibit protein synthesis and non-histones induce RNA synthesis. There are four main steps in the expression of genes. Hence regulation is brought about by the regulation and modification of one or more of these steps. They are
(i) Replication (ii) Transcription
(iii) Processing (iv) Translation
(i) Regulation of replication: Differential gene expression is achieved by gene amplification.
(ii) Regulation of transcription: The regulation of the expression of gene is mainly done at transcription. Hybridization experiments clearly show that production of specialised protein is due to differential gene transcription.
(iii) Regulation of the processing level: Some of the RNA synthesized in the nucleus are destroyed without leaving the nucleus. 80% of the nuclear RNA has no equivalent in the cytoplasm and only 20% if the nuclear RNA is identical in the cytoplasm. All the genes in a cell are transcribed into mRNA at all times, but the mRNA produced by some genes is destroyed rapidly. But the mRNA modeled on other genes are stabilized and only these mRNAs are passed into the cytoplasm.
(iv) Regulation of translation: The control of mRNA-translation is a fundamental phenomenon. In sea-urchin eggs fertilisation is followed by a tremendous increase in protein synthesis; but in the unfertilised egg, there is no protein synthesis. Still the unfertilised egg has complete machinery (i.e., amino acids, ribosomes, mRNA) for protein synthesis. There are two model for regulation in eukaryotes.
(a) Frenster's model: According to 1965, The histones act as repressor's during protein synthesis.
(b) Britten Davidson model: This is also called gene battery model or operon-operator model. It was proposed by Britten and Davidson in 1969. They have been proposed four type of genes namely integrator. sensor, producer and receptor.
Important Tips
(1) Constitutive genes: It constantly express themselves e.g. enzymes of glycolysis, which are also known as house keeping gene, which lacks TATA boxes.
(2)Non constitutive genes: They express themselves only when needed, known as luxury genes Example- Inducible and Repressible genes.
You need to login to perform this action.
You will be redirected in
3 sec
