Some Terms Regarding Enzymes
Category : 11th Class
Zymogens or (Enzyme Precursors) : Certain enzymes are produced by the living cells in an inactive (non-functional) form. They are called the zymogens or proenzymes. It is then converted, usually by proteolysis (hydrolysis of the protein), to the active form when it has reached the site of its activity. Pepsinogen and trypsinogen are zymogens produced by gastric glands and pancreas respectively. They are necessary to life because they degrade dietary proteins into amino acids that are used by the cell.
Pepsinogen is changed to active pepsin by hydrogen ions in the stomach. Trypsinogen is activated to trypsin by an enzyme enterokinase in the small intestine. Once small amount of pepsin or trypsin is formed, it itself catalyzes the activation of remaining proenzyme. This process is called autocatalytic reaction, or autocatalysis.
Isoenzymes (Isozymes) : There are certain enzymes which have slightly different molecular structure but performing the same catalytic function. Such enzymes are called isoenzymes or simply isozymes. Isoenzyme of an enzyme differ from each another in their amino acid sequence, molecular weight, immunological and electrophoretic behaviours. Hence, they can be separated by electrophoresis.
More than 100 enzymes are known to have isoenzyme. A good example of isoenzyme is lactic dehydrogenase (LDH). It catalyzes change of pyruvate to lactate. There are five LDH isoenzymes in muscles of heart. Alcohol dehydrogenase has four isoenzyme in maize. a-amylase (wheat endosperm) has 16 isoenzymes.
Inducible enzymes : An enzyme which is synthesized only in the presence of its substrate (inducer) is called inducible enzyme e.g., \[\beta -\]galactosidase.
Constitutive enzymes (House keeping enzyme) : The enzyme which are found in constant amounts under different growth conditions (regardless of its metabolic states) are called constitutive enzyme e.g., enzymes of sugar breakdown i.e., glycolysis.
Repressible enzymes : The presence of a specific substance may inhibit continued production of specific enzyme (enzyme repressor) e.g., glucokinase.
Ribozymes : Study of post transcriptional processing of RNA molecules has led to the most exciting discovery of the existence of some catalytic RNA molecules which have been called as RNA enzymes or ribozymes. All enzymes are not proteins as confirmed by Cech (1981) and Altman (1983). Ribozyme and RNAase-P are two non protein enzyme where RNA acts as catalyst. Ribozyme was reported from Tetrahymens (a protozoans) by Cech. The substrate for ribozyme is usually an RNA molecule. RNAase-P (Ribonuclease) was discovered by Altman.
Peptidyl transferase is also a non-proteinaceous enzyme, discovered by Noller.
Michaelis constant : Michaelis and Menten (1913) introduced a constant \[{{K}_{m}}\] (Michaelis constant).
It is a mathematical derivative or constant which indicates the substrate concentration at which the chemical reaction catalysed by an enzyme attains half its maximum velocity \[({{V}_{\max }}).\]
\[{{K}_{m}}\]indicates affinity of the enzyme for its substrate.
\[{{K}_{m}}=\frac{1}{2}{{V}_{\max }}\]
\[{{K}_{m}}\]value differs from substrate to substrate because different enzymes differ in their affinity towards different substrates. A high \[{{K}_{m}}\] indicates low affinity while a low \[{{K}_{m}}\] shows strong affinity. Protease acts on different proteins. So it's \[{{K}_{m}}\] value will differ from protein to protein.
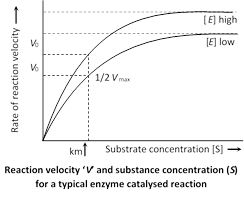
The Michaelis Menten equation description how reaction relatively varies with substrate concentration as given
\[{{V}_{0}}=\frac{{{V}_{\max }}[S]}{{{K}_{m}}+[S]}\]
Where \[{{V}_{0}}\] is the rate of initial reaction; \[{{V}_{\max }}\] is the maximum relative or the reaction rate with excess substrate; \[{{K}_{m}}\] is the Michaelis constant \[={{K}_{2}}+{{K}_{3}}/{{K}_{1}};\text{ }\left[ S \right]\] is the substrate concentration.
The above reaction shows that the greater the affinity between an enzyme and its substrate, the lower the \[{{K}_{m}}\](in units moles per litre) of the enzyme substrate reaction. Stated inversely, \[1/{{K}_{m}}\] is the measure of affinity of the enzyme for its substrate.
Enzyme-inhibitor dissociation constant (Ki) : It is dissociation constant of enzyme\[-\]inhibitor complex.
\[Ki=\frac{[E][I]}{[EI]}\]
Where, E is enzyme and I is concentration of inhibitor.
High Ki decreases enzyme activity while low Ki increases some, it is applicable to competitive inhibitors.
You need to login to perform this action.
You will be redirected in
3 sec
